- Click the cover to access my presentation slides
1. Multilayered omics reveal sex- and depot-dependent adipose progenitor cell heterogeneity
Rana K. Gupta, Yibo Wu
Published in Cell Metabolism
Key words: Development on Existing technology, Proteomics analysis, RNA-seq analysis, Lineage Tracing analysis
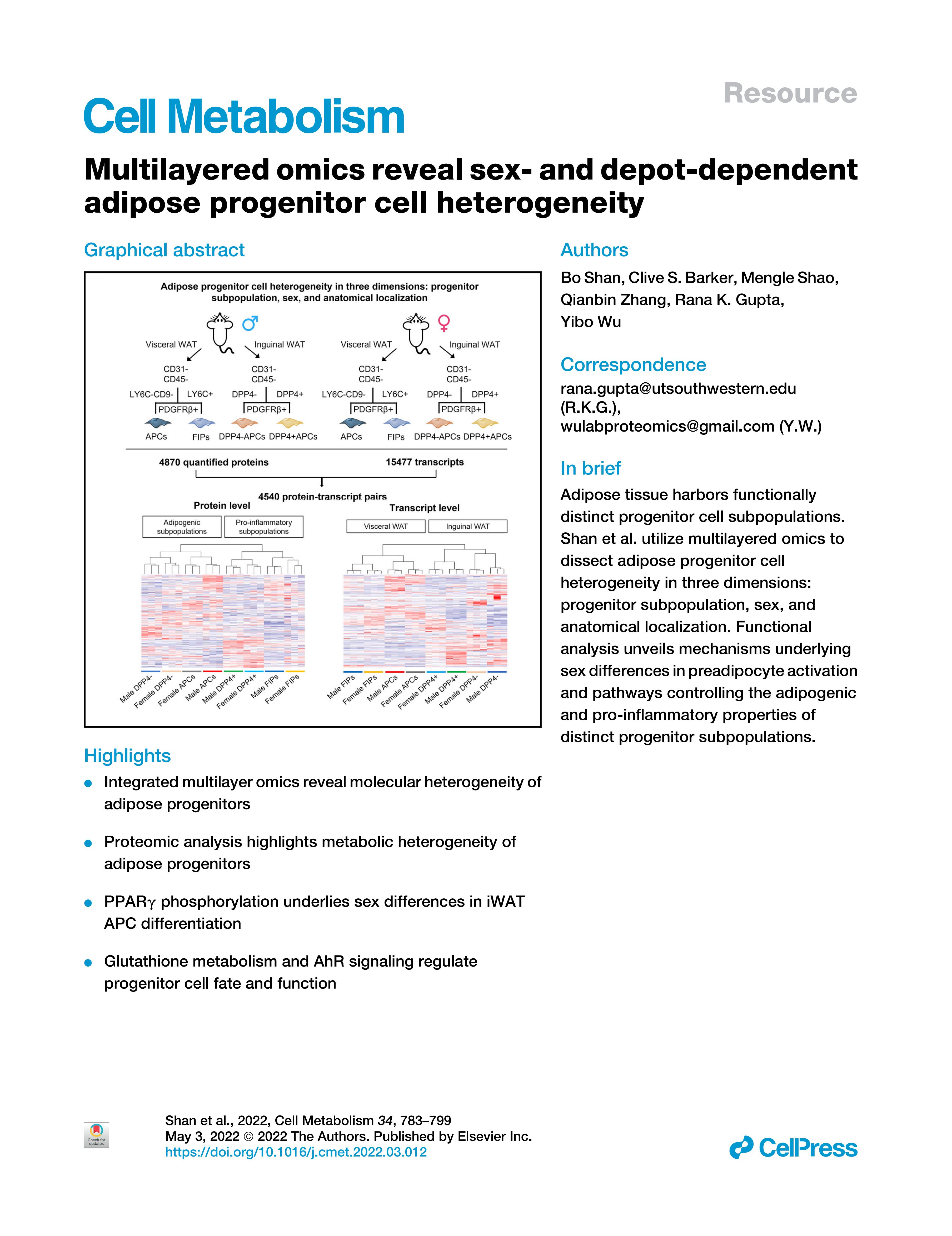
- Comments: The authors of this paper use various methods to analyse the heterogeneity of adipose progenitor cells in mice, and conduct a research focusing on 3 different dimensions: Sex, Location, and Cell type, which largely clarifed the heterogeneity, and demonstrated the power of combining multiple omics in research.
3. Control of osteoblast regeneration by a train of Erk activity waves
Stefano Di Talia, Kenneth D. Poss
Published in Nature
Key words: Breakthrough on the Known phenomena, Regeneration, Erk signaling, Mathematic model

- Comments: This paper employs a mathematical model to simulate the regeneration process and successfully addresses the visualization of Erk signaling in regenerating tissues. Using this model and the visualizing tools, the researchers were able to determine how Erk spreads in zebrafish scales and uncover aspects of the underlying mechanisms of the regeneration process. Most importantly, these methods also offer valuable insights for similar studies in related fields.
7. Mining the CRBN Target Space Redefines Rules for Molecular Glue-induced Neosubstrate Recognition
Sharon Townson, John Castle
Available on BioRxiv
Key words: Development on existing technology, Computional Biology, Molecular Glue, Data Mining

- Comments: The paper predicted over 1,400 CRBN-compatible β-hairpin G-loop proteins across diverse target classes through computational mining of the human proteome using structure-based approaches. It also identified novel mechanisms of neosubstrate recognition. By designing algorithms to screen defined motifs across the entire proteome, the researchers successfully discovered new targets within the CRBN/MGD target space. Furthermore, the paper established a platform that not only broadens the target space but also provides an important reference for similar data mining efforts in this research field.