Benchmarking Algorithms for Batch Correcttion in Single-cell ATAC-seq data Analysis
- Workflow:
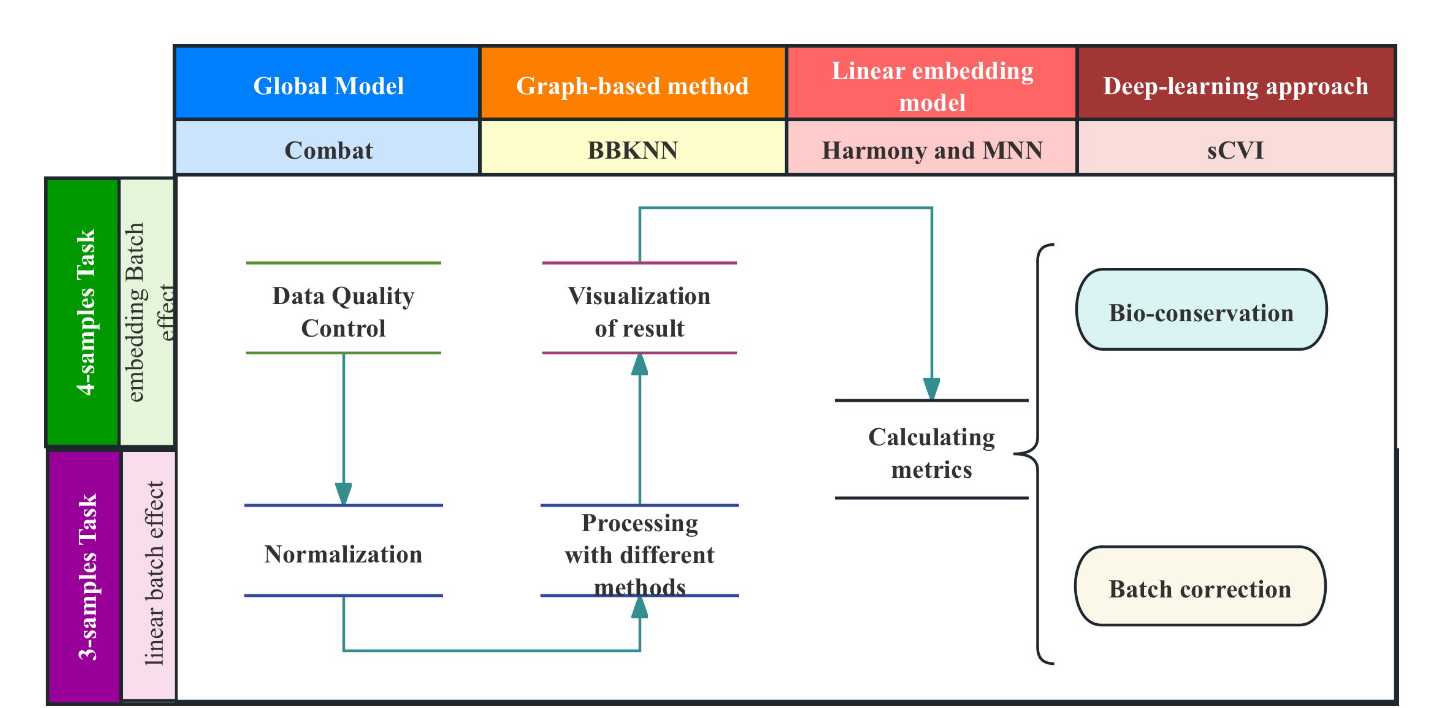
- Abstract:【Purpose】 With the development of high-throughput sequencing technology and single-cell techniques, single-cell chromatin accessibility analysis (Single cell ATAC-seq) has gradually become one of the main means to study the regulatory networks of genome expression. However, in most Single cell ATAC analyses, one of the core issues is how to eliminate unnecessary batch effects caused by different experimental batches. This study aims to compare four batch effect correction algorithms (MNN, Harmony, BBKNN, scVI) in handling two different single-cell ATAC-seq data analysis tasks, to evaluate their performance in preserving biological features and removing batch effects, and to conduct preliminary benchmark testing.【Content】This study first downloads fastqc files of 7 sets of brain single-cell ATAC-seq data published online from the GEO database, and preprocesses the data using software such as Trimgalore, BWA-MEM, SAMTOOL, etc. The obtained Anndata data is then divided into two datasets based on existing batch effects, and four algorithms are used to respectively eliminate batch effects caused by different donors and those caused by different donors and sampling sites. Finally, four metrics are used to evaluate the performance of different algorithms and their preservation effects on biological features, providing references for algorithm selection.【Conclusion】When dealing with datasets with a single source of batch effects, Harmony usually performs excellently in all metrics. When handling datasets with complex batch effects sources, BBKNN performs better. The Harmony and MNN algorithms based on the linear embedding model principle perform outstandingly in different datasets. BBKNN is notably inferior to other algorithms in handling mixed samples. Additionally, the deep learning algorithm scVI also shows stable performance. Through such systematic evaluation and analysis, this study provides a deeper understanding and effective solutions for handling batch effects in single-cell chromatin accessibility data in the field of biomedicine, promoting the development and application of related research, and further revealing the application prospects of deep learning principles in single-cell dataset analysis.
- Key words: Single cell ATAC-seq; Batch effect remval; Benchmark; Single cell data analysis
- link: Read the paper(In Chinese)